Written by Peter Hogg
Neuroscience has become increasingly interdisciplinary, especially as the field enters the era of big data. Confronted with large and complex datasets, scientists need computational tools. But traditional training in the life sciences often leaves scientists with a gap in understanding how to develop or use some of these computational tools. During the COVID-19 lockdown, Databinge was relaunched to overcome this gap.
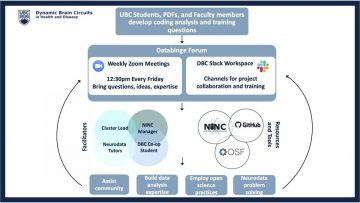
Databinge is a weekly virtual meeting hosted by the Dynamic Brain Circuits in Health and Disease research cluster of the DMCBH. The purpose of the meeting is for members of the UBC neuroscience community to come together and discuss data analysis problems. In these virtual and casual meetings, people come with their questions and problems, so Neurodata Tutors can help solve the problems using programming. The meetings start with large group sessions where anyone can talk about what they are working on and ask for input to solve their analysis problems. These large group discussions are excellent for brainstorming and crowdsourcing solutions, especially when there might be existing solutions and tools in the literature or ones developed by group members. During a Databinge meeting, people are also matched with 1-2 Neurodata Tutors that have expertise in solving a specific problem. These tutors come from a range of backgrounds. They are not only experienced in using various popular programming languages, such as Matlab, Python and R, but also knowledgeable in common analytic approaches such as image analysis, statistics and machine learning. In smaller breakout meetings, the Neurodata Tutors get into the specifics of how to implement solutions and teach the community how to automate data analysis using computer programming.
Databinge has helped multiple projects move forward by fostering collaboration between the Neurodata Tutors and members of the UBC neuroscience community. These projects span over several domains of neuroscience, from molecular biology to animal behavior. One successful example was developing a pipeline to analyze calcium imaging data of neurons from a Huntington’s disease mouse model. The data videos were originally analyzed by hand, where the researcher manually selected a region of interest from videos to generate calcium traces. This process was labour intensive and took too much time. By incorporating codes to select regions of interest and extract calcium signals, the new analysis pipeline now allows for automatic detection and quantification of calcium signals. The resulting pipelined simplified the researcher’s analysis and saved them a significant amount of time by fully automizing the process.
Another successful collaboration was solving a data visualization problem regarding the expression of palmitoyltransferases. With increasingly large and complicated RNA expression datasets, there are challenges for displaying all information rationally and logically. The solution was to develop an interactive heatmap written in a combination of Python and Javascript, so that researchers could select subsets of gene s and brain regions, and examine this smaller sample of the overall dataset. Simpler projects also include writing scripts to automatically convert from one file format to another or writing ImageJ/Fiji macros. Ongoing projects include developing tools for behavior quantification in mice from pose estimation data generated using the popular machine learning tool DeepLabCut.
Anyone can visit and participate in the Databinge meetings via the shared Zoom link on the DMCBH website (link). We believe everyone offers unique perspectives and insights into data analysis problems.


